My Project - FASTA Analyser V1
Getting back to the project I gave myself, here’s the first piece of code (not following tutorials) I had a go at writing.
The aim here was to allow a user to enter a DNA sequence in text format, for example ‘CGATCGATCGAAATCGATGCCTAATCCATAGAGGGGA’ and to have Python return the length, G, C, A and T content. Nothing too crazy or complicated, but I thought this would be a nice place to start. I tried a few different methods to achieve this, but here’s the code I settled with.

OK, so let’s break down what I’ve written here. Looking at the code you might be asking what the section at the top in triple quotation marks is for?
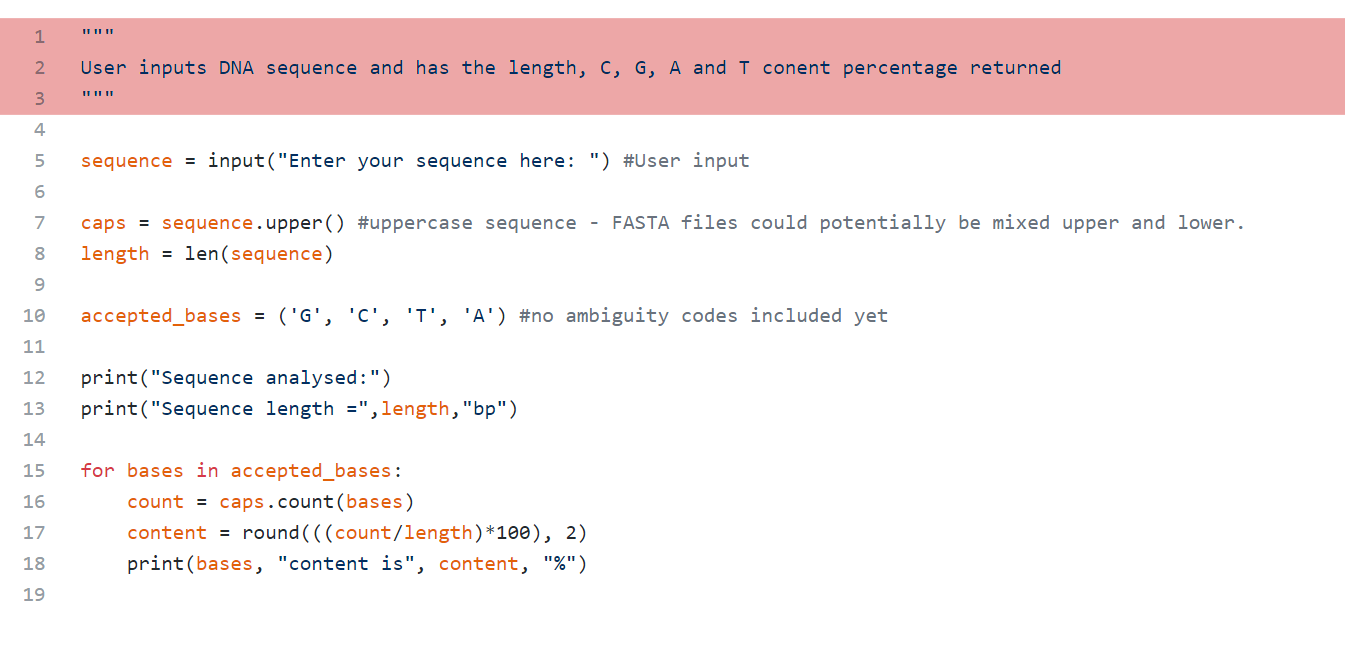
This is called a docstring and should be written into methods and modules using the syntax seen here. Essentially it’s a description of what the code does and comes in handy when other people adopt your code into their programs. For example, if I saved this code as fastaanalyser.py, and somebody imported this into their program, they can use Python’s help function to access the docstring and find out what the module does. In this example, this would be done simply by using ‘help(fastaanalyser)’ and Python would return the string User inputs DNA sequence and has the length, C, G, A and T content percentage returned. I definitely was not writing docstrings when I first played around with this code, but including them can save a lot of time for future users.
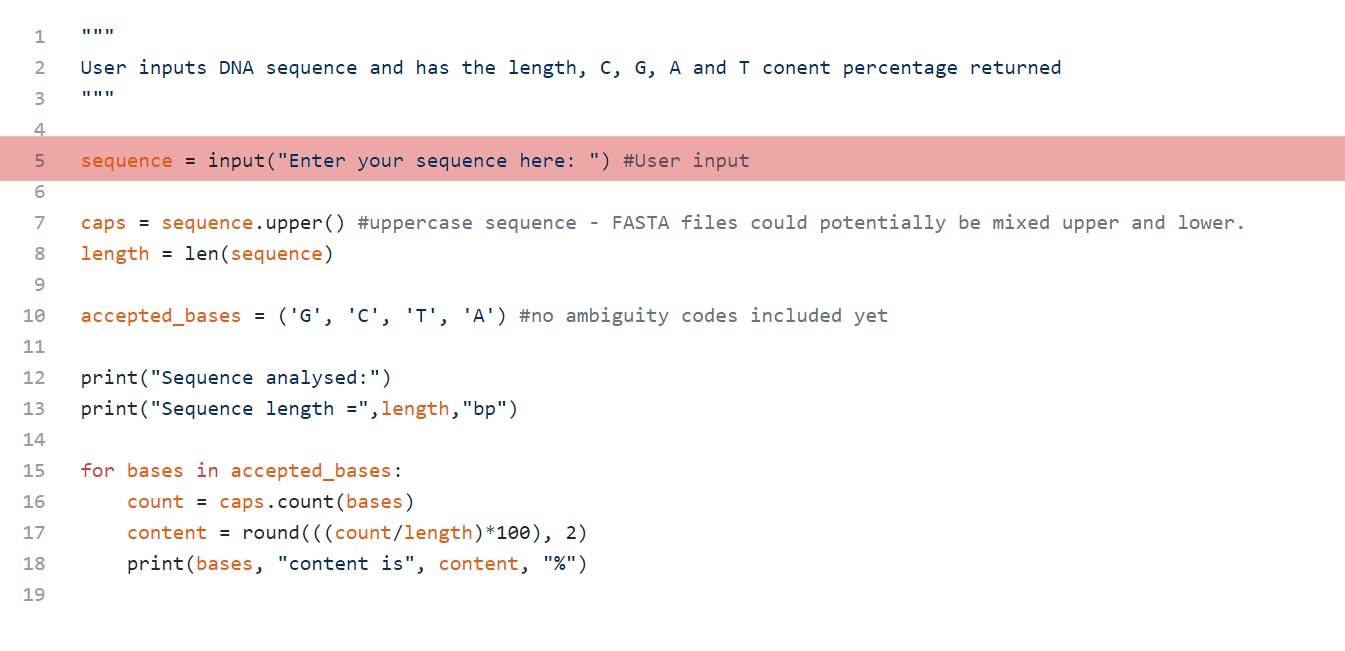
The next section is where the user will be prompted for input. This is really to achieve in Python and the syntax can be seen here. There are a couple of different input methods that can be used, but this one processes the user’s input as a string. I will do a post on the other input methods at some point.
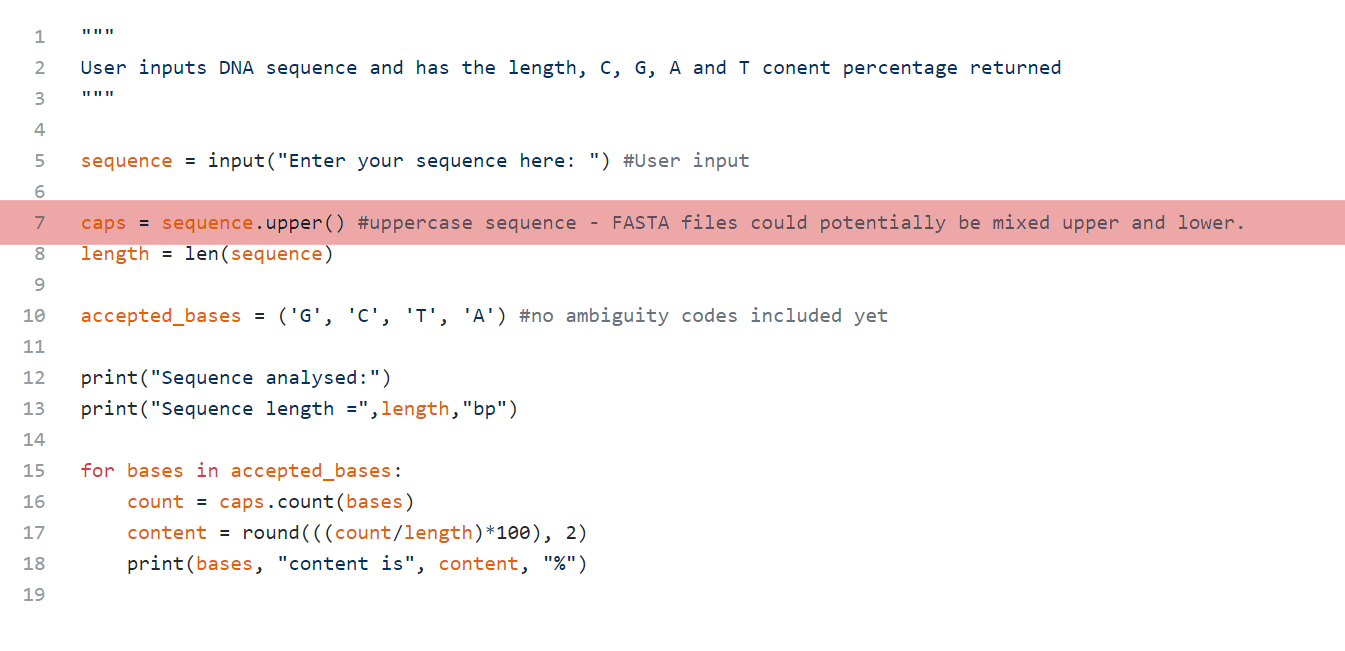
First, the script takes the user’s input and changes is all to uppercase. This is done use the ‘upper’ method seen here. I included this as FASTA sequences can sometimes be a mix of upper and lower case. Making all the characters uppercase saves me from having to include the lower case letter in the accepted bases tuple. I used a tuple for the accepted bases as this list will not be altered during the running of this code. Tuples, unlike lists and dictionaries are unmutable and cannot be changed.

Here we see Python’s length method. This will return the length of the user’s sequence as a variable called ‘length’. This variable ‘length’ is then printed to the screen.

Highlighted here is the main working of the analyser. To reduce the amount of code required, I have used a for loop. The code block within the for loop counts the number of times a base appears, calculates that as a percentage over the entire sequence and then prints the information to the screen. The for loop is set up so that for each base in ‘accepted_bases’, the code will be run once. So this analyser will start off by counting the number of ‘G’s, working that out as a percentage then printing the result. It will then proceed to do the same again for ‘C’, ‘T’ and ‘A’.
Here’s what the result looks like when using the sequence example given at the top of this post.
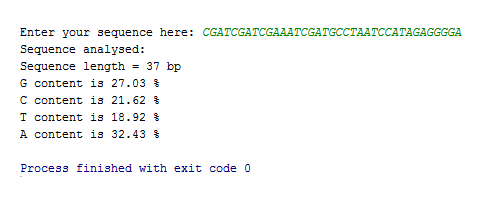
There are a few things I could do to improve this code. For example, currently it has no way of catching any errors. A user could enter an integer and Python would happily execute the code - producing no result obviously. A try and except block could be added to make sure the user inputs a string. Another improvement could be to user some aspects of the Tkinter module, allowing the user to select a file directly, instead of having to copy and paste a sequence into the interpreter.
I will re-visit this code and add these features in at a later date and talk about it in another post. For now, this code works nicely and was a great way to try out some of the concepts I’d learnt in a real situation.
